Getting Started¶
Univariate Functions¶
The standard workflow for autodiff is to first initiate a
Variable, or several Variables. We then use these Variable
to construct Expressions, which can then be queried for values and
derivatives.
In [24]:
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from autodiff.forward import *
Suppose we want to calculate the derivatives of
\(f(x) = \cos(\pi x)\exp(-x^2)\). We can start with creating a
Variable called x.
In [3]:
x = Variable()
We then create the Expression for \(f(x)\). Note that here
cos and exp are library functions from autrodiff.
In [4]:
f = cos(np.pi*x)*exp(-x**2)
We can then evaluate \(f(x)\)‘s value and derivative by calling the
evaluation_at method and the derivative_at method. For
derivative_at method, the first argument specifies which variable to
take derivative with respect to, the second argument specifies which
point in the domain are the derivative to be calculated.
In [5]:
f.evaluation_at({x: 1})
Out[5]:
-0.36787944117144233
In [6]:
f.derivative_at(x, {x: 1})
Out[6]:
0.7357588823428846
The derivative_at method supports second order derivative. If we
want to calculate \(\dfrac{d^2 f}{d x^2}\), we can add another
argument order=2.
In [7]:
f.derivative_at(x, {x: 1}, order=2)
Out[7]:
2.895065669313077
Both the methods evaluation_at and derivative_at are vectorized,
and instead of pass in a scalar value, we can pass in a numpy.array,
and the output will be f’s value / derivative at all entried of the
input. For example, we can calculate the value, first order derivative
and second order derivative of \(f(x)\) on the interval
\([-2, 2]\) simply by
In [8]:
interval = np.linspace(-2, 2, 200)
values = f.evaluation_at( {x: interval})
der1st = f.derivative_at(x, {x: interval})
der2nd = f.derivative_at(x, {x: interval}, order=2)
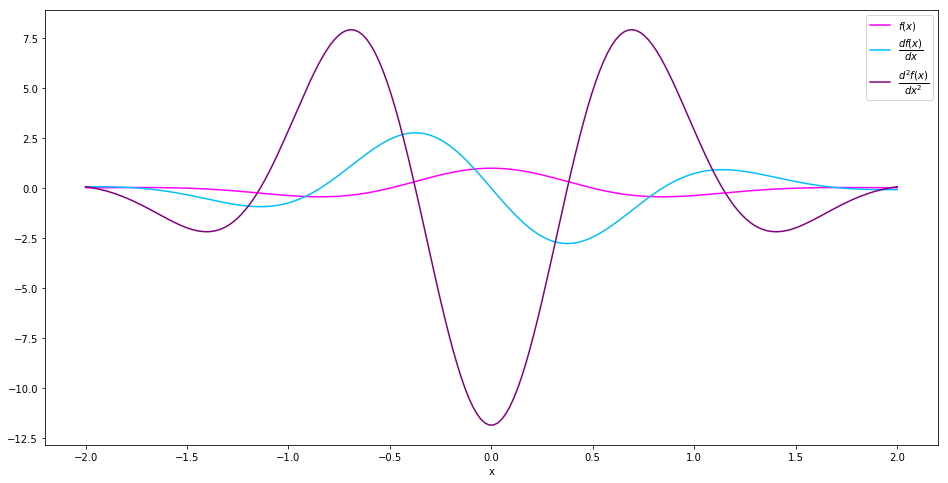
Let’s see what they look like.
In [9]:
fig = plt.figure(figsize=(16, 8))
plt.plot(interval, values, c='magenta', label='$f(x)$')
plt.plot(interval, der1st, c='deepskyblue', label='$\dfrac{df(x)}{dx}$')
plt.plot(interval, der2nd, c='purple', label='$\dfrac{d^2f(x)}{dx^2}$')
plt.xlabel('x')
plt.legend()
plt.show()
Multivariate Functions¶
The workflow with multivariate functions are essentially the same.
Suppose we want to calculate the derivatives of
\(g(x, y) = \cos(\pi x)\cos(\pi y)\exp(-x^2-y^2)\). We can start
with adding another Variable called y.
In [10]:
y = Variable()
We then create the Expression for \(g(x, y)\).
In [11]:
g = cos(np.pi*x) * cos(np.pi*y) * exp(-x**2-y**2)
We can then evaluate \(f(x)\)‘s value and derivative by calling the
evaluation_at method and the derivative_at method, as usual.
In [12]:
g.evaluation_at({x: 1.0, y: 1.0})
Out[12]:
0.1353352832366127
In [13]:
g.derivative_at(x, {x: 1.0, y: 1.0})
Out[13]:
-0.27067056647322535
In [14]:
g.derivative_at(x, {x: 1.0, y: 1.0})
Out[14]:
-1.0650351405815222
Now we have two variables, we may want to calculate
\(\dfrac{\partial^2 g}{\partial x \partial y}\). We can just replace
the first argument of derivative_at to a tuple (x, y). In this
case the third argument order=2 can be omitted, because the
Expression can infer from the first argument that we are looking for
a second order derivative.
In [15]:
g.derivative_at((x, y), {x: 1.0, y: 1.0})
Out[15]:
0.5413411329464506
We can also ask g for its Hessian matrix. A numpy.array will be
returned.
In [29]:
g.hessian_at({x: 1.0, y:1.0})
Out[29]:
array([[-1.06503514, 0.54134113],
[ 0.54134113, -1.06503514]])
Since the evaluation_at method and derivarive_at method are
vectorized, we can as well pass in a mesh grid, and the output will be a
grid of the same shape. For example, we can calculate the value, first
order derivative and second order derivative of f(x)f(x) on the interval
\(x\in[−2,2], y\in[-2,2]\) simply by
In [20]:
us, vs = np.linspace(-2, 2, 200), np.linspace(-2, 2, 200)
uu, vv = np.meshgrid(us, vs)
In [21]:
values = g.evaluation_at( {x: uu, y:vv})
der1st = g.derivative_at(x, {x: uu, y:vv})
der2nd = g.derivative_at((x, y), {x: uu, y:vv})
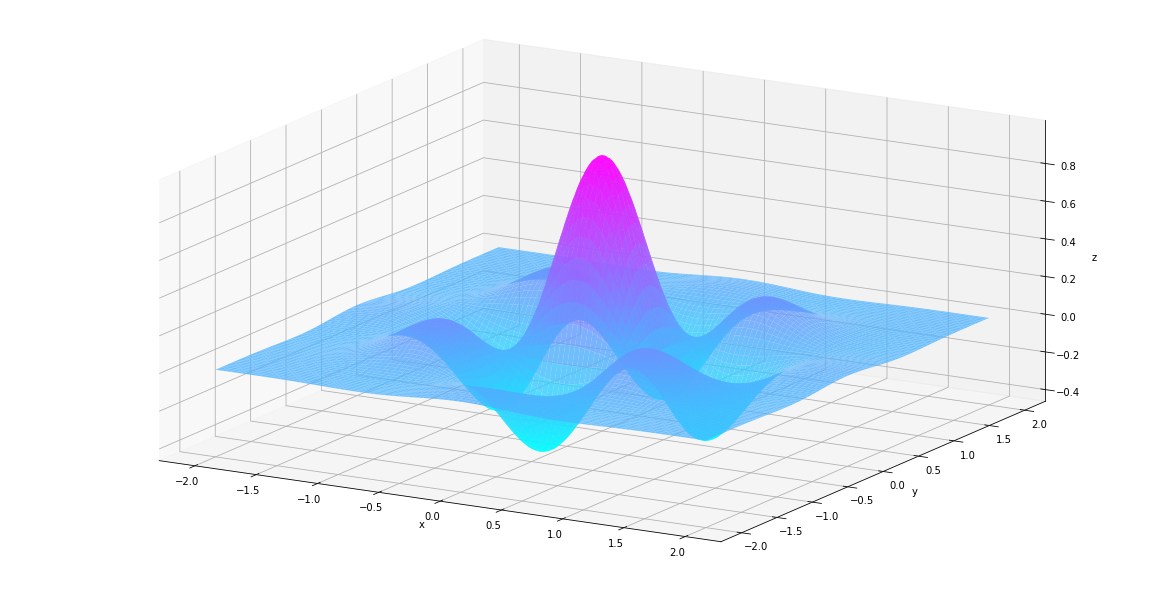
Let’s see what they look like.
In [22]:
def plt_surf(uu, vv, zz):
fig = plt.figure(figsize=(16, 8))
ax = Axes3D(fig)
surf = ax.plot_surface(uu, vv, zz, rstride=2, cstride=2, alpha=0.8, cmap='cool')
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_zlabel('z')
ax.set_proj_type('ortho')
plt.show()
In [25]:
plt_surf(uu, vv, values)
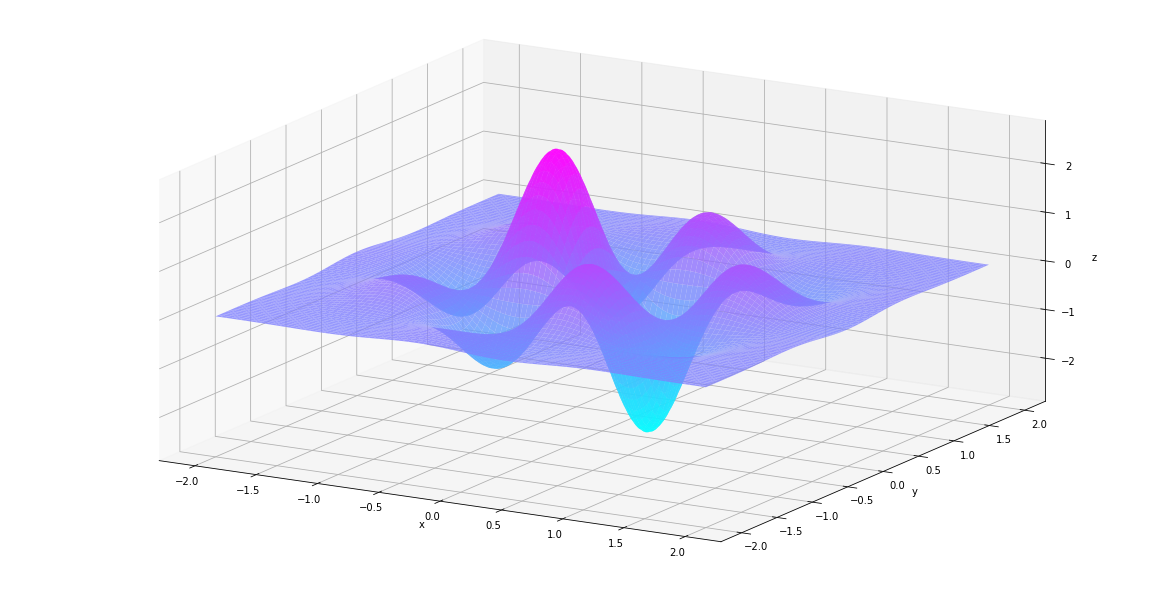
In [26]:
plt_surf(uu, vv, der1st)
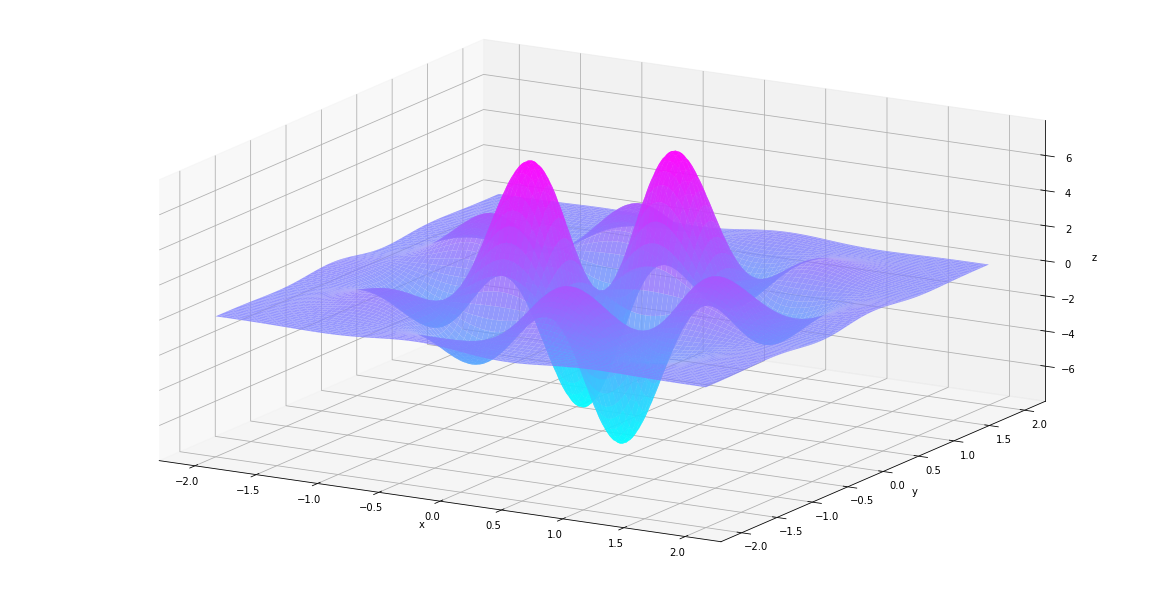
In [27]:
plt_surf(uu, vv, der2nd)
Vector Functions¶
Functions defined on \(\mathbb{R}^n \mapsto \mathbb{R}^m\) are also
supported. Here we create an VectorFunction that represents
\(h(\begin{bmatrix}x\\y\end{bmatrix}) = \begin{bmatrix}f(x)\\g(x, y)\end{bmatrix}\).
In [30]:
h = VectorFunction(exprlist=[f, g])
We can then evaluates \(h(\begin{bmatrix}x\\y\end{bmatrix})\)‘s
value and gradient
(\(\begin{bmatrix}\dfrac{\partial f}{\partial x}\\\dfrac{\partial g}{\partial x}\end{bmatrix}\)
and
\(\begin{bmatrix}\dfrac{\partial f}{\partial y}\\\dfrac{\partial g}{\partial y}\end{bmatrix}\))
by calling its evaluation_at method and gradient_at method. The
jacobian_at function returns the Jacobian matrix
(\(\begin{bmatrix}\dfrac{\partial f}{\partial x} & \dfrac{\partial f}{\partial y} \\ \dfrac{\partial g}{\partial x} & \dfrac{\partial g}{\partial y} \end{bmatrix}\)).
In [31]:
h.evaluation_at({x: 1.0, y: -1.0})
Out[31]:
array([-0.36787944, 0.13533528])
In [35]:
h.gradient_at(0, {x: 1.0, y: -1.0})
Out[35]:
array([0., 0.])
In [33]:
h.jacobian_at({x: 1.0, y: -1.0})
Out[33]:
array([[ 0.73575888, 0. ],
[-0.27067057, 0.27067057]])